iPaulina
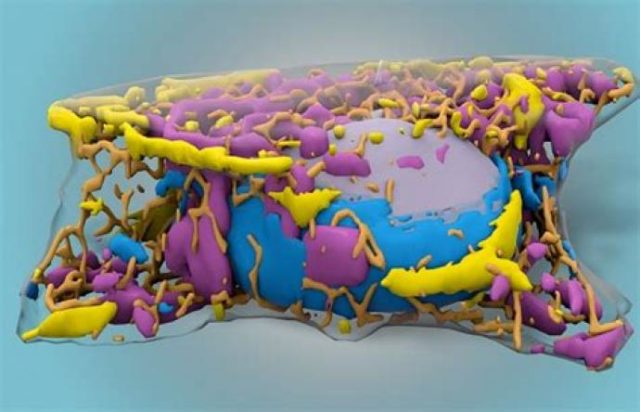
Machine learning and microscopy
The benefits of using AI in researchWith recent advances in machine learning (ML), an increasing number of scientists have been implementing ML techniques in image-based studies in biology and medicine, with the growing complexity of microscopic images providing an opportunity for computer aid in these fields. Machine learning helps speed up tasks, and with how time-consuming it can be to take information from images at different scales, all with different morphology and levels of noise, utilising algorithms can help automatically detect patterns in images and cells themselves.
Machine learning is a subset of artificial intelligence in which an algorithm learns from data and finds patterns and relationships. When observing cells, researchers typically use fluorescent tagging or stains to identify characteristics of a cell. In 2018, using a computer model to focus on details without using markers, an algorithm was able to correctly differentiate between smaller structures. To create an algorithm with high accuracy, a large amount of data is required to train models, and this had to be done with a lot of manual annotation. This helped with developing a neural network that could distinguish between cell types, label them down to where neurons’ bodies ended and their axons and dendrites began, and distinguish dead cells from living ones.
In addition to labelling, another area in which implementing ML models has been proven useful is in removing noise from microscopic images. Noise is caused by insufficient light, causing the graininess we often see in photographs taken at night. There will always be noise in photographs and some cannot be avoided. However, although algorithms to remove noise have been used for years, using deep learning models produces results that are far more effective
Self-supervised learning is where the network is forced to learn what data has been withheld in order to solve a problem. Normally, with supervised learning, the machine knows what is looking for because it has been trained with images that have clean versions. However with self-supervised learning, researchers have to try using algorithms that train themselves. The outputs from these models look good, but that does not mean they are real.
The primary concern when using models is often whether data is being changed and the number of mistakes being made. With the labelling models, biologists consistently check computers’ work to ensure that structures have been defined accurately. In these scenarios, the original image is not changed, but another layer is added on top. With de-noising algorithms, the image has been modified to produce a cleaner result. If the noise in the original image is stronger, then these changes will be more pronounced. In one project, researchers were attempting to remove blur but rather than doing so, the algorithm simply picked up the pattern of stripes in the original images and removed any stripes in new images it had to de-noise, although this was fixed by adding training data.
There are now an increasing number of repositories containing different algorithms by researchers to de-noise images and in other areas like segmentation and classification. Neural networks are now being used in mapping brain tumours, studying RNA localisation and electron microscopy. As these models continue to develop in complexity and accuracy, and researchers continue to share results and work on algorithms together, we can already see how beneficial implementing AI in research can be.
Radhika VI